The real title of the paper is “Multiple self-splicing introns in the 16S rRNA genes of giant sulfur bacteria”. But who’s going to fall out of their chair for that?
The truth is, we do have aliens peppered among us. Think about Men in Black: plenty of space creatures, but Homo sapiens remains completely oblivious. Will Smith can separate the aliens from the humans (and look super smokin’ in the process), but that’s only because he knows what he’s looking for. But scientists…often we don’t know what we should be looking for.
Since we started using DNA to find species in natural environments, we’ve built up a fairly good idea of what we can usually expect to find in different habitats. Of course, there is a HUGE amount of undiscovered biodiversity, and many novel creatures yet to be discovered, but we at least know the major players. Bacteria, Archaea, Eukaryotes, and Viruses–we group species into these broad taxonomic groups since they represent the major domains of life on Earth (Well, so far at least…).
Our molecular toolkit is pretty badass. Us scientists can use the Polymerase Chain Reaction to amplify conserved, universal genes like those encoding the ribosomal subunits. Most living things can’t function without ribosomes, so their genomes must contain the instructions. Just like a handheld supermarket scanner, we can go into any environment (any store) and follow simple steps to get a list of ribosome DNA sequences and match them to species (akin to scanning the barcodes to pull up information about the products populating the shelves).
The problem? This approach assumes that all barcodes adhere to a standard format. Which normally, they do:

Except sometimes, they look like this:

I’m not an idiot–I know both of these are obviously barcodes. Our human eyes can see that. But the supermarket scanner can’t. Haven’t you ever been in line at the store (in a massive hurry), where you grab an item off the shelf and rush over to the cashier in hopes of quickly paying (to avoid being too fashionably late to your fabulous gala ball, of course)? That’s where things go awry and your item won’t scan, so you’re forced to wait (impatiently tapping your feet, dreaming of champagne) while another employee is summoned to fetch the correct price.
Everyone has faced this problem at the supermarket. Sure, the barcode on your idem looked like every other barcode, but when you go to scan it, the system can’t find the ID. If you knew it would be a problem ahead of time, you would have come up with another way for the cashier to access the needed information–in this case, you may have taken a picture on your phone to show the item on the shelf next to the price. If the mechanisms we have in place just can’t read a barcode (at the supermarket or in the lab), there must be an alternate solution to link the necessary information. Barcodes are meant to save us time, after all.
In a retail setting, a rogue barcode is not a big deal. There are plenty of bored teenagers who can be summoned to do the legwork. But in science, tricky barcodes are a more worrying issue. When applying DNA tools to study natural environments, we use molecular information to gather information about the community at hand. Each DNA barcode is useful because it returns a taxonomic name, which we can use to infer what a species might be doing–what its eats, the nutrients or chemicals it might help to break down, or the other species it might prey upon. When we’re delving into natural environments using DNA tools, we’re essentially scanning for barcodes in the dark and assuming we’re catching everything. And we certainly assume we’re capturing the most abundant and ecologically important species.
Except…the study I first mentioned puts a damper on this rose-tinted view. Turns out that some bacteria–giant sulfur bacteria, to be exact–actually have pretty f%&*#ing uncooperative barcodes. Because of the presence of noncoding introns within gene sequences, their ribosomal barcodes are longer than we’re normally used to seeing: we can’t scan them using our standard lab protocols. Salman et al. (2012) suggests that our inability to detect barcodes from sulfur bacteria may be seriously skewing our DNA-based interpretations of how ocean ecosystems function.
Large sulfur bacteria cycle nutrients LIKE A BOSS. They’re ecological powerhouses, involved in the turnover of sulfur, nitrogen and phosphorus in seafloor habitats. So this barcode issue is a big deal–if we really want to understand ecosystem function, we definitely need to be able to spot these giant sulfur bacteria when they’re present (especially since they tend to occur at high densities). If we haven’t been finding these species using our bog standard molecular approaches, we might be totally misinterpreting species assemblages in sediment communities, and consequently, the process of nutrient cycling in the oceans.
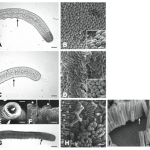
The sulfur bacteria who have rogue barcodes are pretty awesome species, too! Included in this group is the largest known bacteria, Thiomargarita namibiensis. It quite literally is a giant bacteria, measuring in at 0.1mm to 0.75 mm.
The species name namibiensis indicates its origins in coastal Namibia, meaning Sulfur pearl of Namibia. This is a reference to the fact that the bacterium-chains have the appearance of a thin string of pearls, due to microscopic sulfur granules inside the bacteria, reflecting the incident light. [Wikipedia]
Wikipedia says this species looks like pearls, but I think they look like anal beads:

T. namibiensis lives in ocean mud infused with sulfur, its favorite nutrient to gobble up. The species’ ability to oxidize sulfur (often in low oxygen conditions) makes them the Betty Ford clinic of the deep-sea: they intervene to remove otherwise toxic chemicals from the environment, using their specially adapted metabolic pathways.
Thiomargarita namibiensis…oxidize nitrate into sulfide in the low-nitrate conditions of their oxygen-poor habitat. This bacterium has accomplished this by having the ability to store both sulfur and nitrate. Each individual bacterial cell is almost entirely made up of a liquid vacuole, in which is contained large amounts of nitrate, often 10,000 times the amount in the surrounding seawater. The bacteria use this store of nitrate, gathered when it is abundant in the water surrounding the cells, to oxidize the sulfur from the seafloor. The cytoplasm of the cells line the inside of the cell membrane, allowing the bacterium the flexibility to diffuse the nitrate with the outside sulfide and giving it its huge size. Unlike its relation Triploca, Thiomargarita namibiensis does not have to migrate vertically in order to collect enough nitrate to properly oxidize; its cells can live submerged in the sediment for up to three months before requiring nitrate from the outside environment. They have also developed capabilities to withstand extreme environmental changes, such as an influx of oxygen or sulfide. [Microbe Wiki]
Sulfur bacteria may have been escaping our molecular scrutiny, but now the secret is out. Given their “alien” ribosomal sequences, we can design new methods to capture the enigmatic barcodes of these cool giant microbes. But science carries on, and there are plenty more questions to ask:

Reference: Salman, V. et al. (2012). Multiple self-splicing introns in the 16S rRNA genes of giant sulfur bacteria. Proceedings of the National Academy of Sciences, USA, 109(11):4203-4208.