A “species” is a hypothesis. And for microscopic critters, this hypothesis is very often wrong.
Everyone knows I despise charismatic megafauna (especially dolphins). I will now secretly admit that I also don’t care much for charismatic invertebrates. I mean, Yeti crabs are pretty much the Lindsay Lohan of marine creatures – they’re just too damn attention-seeking with their bristly claws and all. And they’re everywhere…
What I do love are cryptic species – nondescript, gooey specimens that all look the same, but have surprisingly distinct genomes. Candlelight and soul forever, I dream of myself and them together!

Looking at the DNA of microscopic animals is full of mystery and intrigue – it feels a bit like solving a crime (albeit a PG-13, Nancy Drew type of crime). Traditional taxonomy just doesn’t have that thill for me – it is slow and steady, requires hours under the microscope, and has an unsettling degree of subjectivity. DNA, on the other hand, is extremely objective – you can’t argue* with good data sequenced from a trustworthy spot in the genome. Those ATCGs just don’t lie.
But that doesn’t mean DNA is a magic bullet. DNA provides a new type of evidence for making decisions about species, but that evidence still has to be robustly analyzed and interpreted in the context of historical knowledge (taxonomic classifications and anatomical features of specimens).
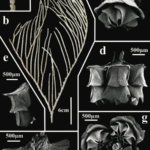
A recent paper by Jörger et al. (2012) displays the power of DNA to differentiate amongst gelatinous blobs. The authors analyzed molecular evidence alongside body features to investigate sea slug species in the genus Pontohedyle, a group which taxonomists thought they had described LIKE A BOSS. Anatomy didn’t tell much of an exiting story – specimens could be lumped into two different groups based on appearance, and both these species were presumed to live pretty much anywhere in the ocean. [Sidenote: this is a pretty common assumption for microscopic animals and is known as the “meiofaunal paradox”. Victorian scientists would think “Oh, they all look the same, so lets describe these as the same species.” because they didn’t have any badass knowledge about DNA, and they didn’t have a time machine. But nowadays, this thinking is in direct conflict with our knowledge about these species–microscopic animals have been shown to have limited ability to move around, even during larval stages. They pretty much get stuck in the marine neighborhood where they grew up, so how can they spread their sweet DNA all around the world?]
The researchers started to look a little bit closer, baby, and study how sea slugs get it on, get it on. Because…
Uncovering putative cryptic lineages is fundamental not only for our advances in understanding speciation processes in meiofaunal taxa [microscopic multicellular animals <1mm long, such as nematodes, copepods, and tardigrades], but also to understanding historical biogeography.
Jörger et al. analyzed the sea slug DNA in every way they could think of. First off, they looked at the story from three different genes: Cytocrhome c Oxidase subunit 1 (the “barcoding” locus in the mitochondrial genome), 16S rRNA (a gene encoding a ribosome subunit in the mitochondria), and 28S rRNA (a ribosome subunit gene in the nuclear genome).
All three genes told the same story. The results of different data analysis methods were consistent. And wow, they sure threw all the data analysis acronyms they could think of at that DNA:
…we apply four independent methods of molecular based species delineation: General Mixed Yule Coalescent model (GMYC), statistical parsimony, Bayesian Species Delineation (BPP) and Automatic Barcode Gap Discovery (ABGD). The secondary species hypothesis (SSH) is gained by relying only on uncontradicted results of the different approaches (‘minimum consensus approach’).
In the end, 2 morphological species became 12 cryptic species (each color in the this tree below representing a different cryptic species):
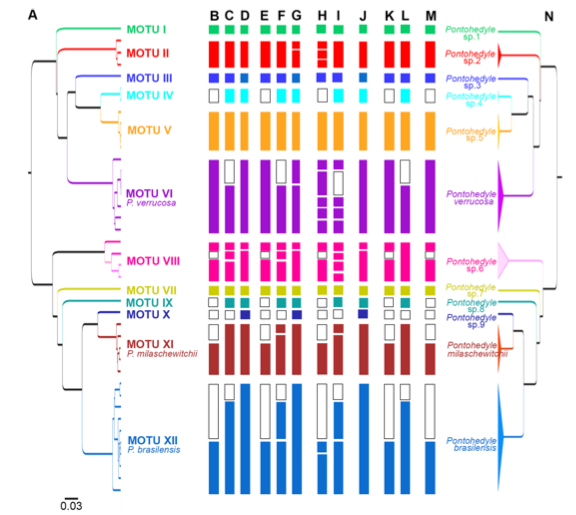
The power of antatomical features was, er, not so powerful after all.
Previously used external morphological characters such as the shape of oral tentacles, body length and width, or color of the digestive gland (e.g., [68]) depend highly on the stage of contraction and nutrition, and are variable through time for each individual [40,41] and therefore inappropriate for species delineation.
The color of the digestive gland?! Really, guys?
So think about what this means for our understanding of biodiversity on earth. This is just one study, from one genus out of thousands in the taxonomic classification of animals (and there’s still a lot of microscopic critters we haven’t even described). Evidence like this comes out all the time. For example, a study focused on the nematode Pellioditis marina (a well known estuarine species living in Northern Europe) found that this “species” is actually a complex of at least 4 genetically distinct cryptic species. Just when we think we’re making good progress on taxonomy, DNA is routinely telling us that we probably have to split every known microbial eukaryote species into 3-4 cryptic entities.
And on top of that, Jörger et al. remind us of how little of the earth’s surface we’ve actually sampled:
Although we present the first study on meiofaunal slugs with representative worldwide taxon sampling, we know our dataset is likely to represent only a fragment of the hidden diversity in the genus because a) tropical sands still are largely unsampled, b) suitable habitats are patchy and disjunct, and c) the indication of accumulated diversity in geographically small areas (e.g., three distinct genetic Pontohedyle lineages on the island of Moorea).
Scientists–and humans–are on an eternal quest to talk about species. It helps us organize our knowledge of the world, and helps us to understand historical processes which have influenced how things evolve and how they are distributed geographically. There’s always a lot of press surrounding the latest paper claiming the official and absolute estimate of the number of species on earth (here’s one hyped up example). Personally, I don’t take much stock in these, because I don’t think we have nearly enough information to even make ballpark guesses. We have NO genetic information about most microscopic animals on earth. Hell, the Guinea Worm can’t even get its genome sequenced. For small non-parasitic wormy things, we’re just lumping similar-looking blobs into categories after a quick peek down the microscope. Its like saying all blonde haired people belong to one species, and all redheads belong to another – a.k.a, surface appearances take precedence, end of story.
Next time you hear a “global species estimate”, don’t say you believe it, please don’t say you believe it!
*Ok, well you can argue, and people do. But that is another blog post for another day. Just work with me here, people.
Reference:
Jörger KM, Norenburg JL, Wilson NG, Schrödl M. (2012) Barcoding against a paradox? Combined molecular species delineations reveal multiple cryptic lineages in elusive meiofaunal sea slugs. BMC Evol Biol,12(1):245.
Share the post "When 2 becomes 12: Cryptic species need some love like they’ve never needed love before"


And so we find more and more species at the same time as the concept “species” looses all practical meaning. We cannot identify them without chemicals. We no longer know what biodiversity means. We don’t know how to value a species. Just as well we could dump the term and go on with genomes only. Imagine the joy experienced on a Sunday-afternoon walk: I have seen 253 different butterfly genomes!
We are running into a scientific and philosophic dead end.