A professor once told me that if you removed everything from earth and just left the nematodes you would still recognize the outlines of everything. I have absolutely no idea if this is even remotely true. I do know that, hyperbole aside, nematodes represent one of the most abundant forms of life on earth. The phyla represent well over 28,000 described species of freshwater, marine, terrestrial, and parasitic round worms. On the seafloor they account for 85-95% of the organisms.
How many nematodes species are there out there unknown to science? Some estimate upwards of 1,000,000. However, authors have placed the undiscovered deep-sea nematode diversity at 1,000,000 by itself. Of course these extrapolations are fraught with uncertainty and more realistic assumptions of deep-sea nematode diversity are nearer to 10,000.
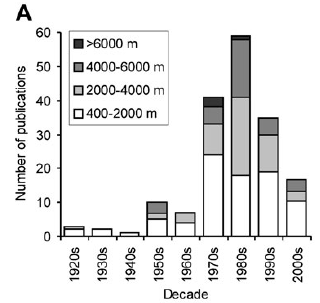
If we assume that 10,000 is the cap for global nematode diversity, how many species do we have to still describe? About 9400. In a recent study, a group of nematode biologists conducted a comprehensive review of the literature and discovered that 638 valid species of deep-sea nematodes belonging to 175 genera and 44 families are described.
However, many, many more are waiting description whether you believe the 10,000 or 1,000,000 estimate. Consider this all we know about all deep-sea nematodes is based on a meager 473 samples that cover a bottom area of 60-70 meters square, about the area covered by a small house.
What is alarming is not the meager amount of deep-sea sampled, although a concerted effort is needed for exploration, but rather that the number of papers describing new species is actually half what it was decade ago and a third of decade before that.
With technological advancements and continued exploration of the deep, why is the description of new species not increasing?
Because fewer and fewer scientists are interested in describing species.
The biological sciences have devalued taxonomy as field and as such few positions are available for doing this kind of work. We have entered a positive feedback loop, where the end result is that we are actually retarding science. Species are going extinct before we can describe them and ultimately we are culpable.
We are ass deep in a biodiversity crisis and we are getting it in both ends.
Miljutin, D., Gad, G., Miljutina, M., Mokievsky, V., Fonseca-Genevois, V., & Esteves, A. (2010). The state of knowledge on deep-sea nematode taxonomy: how many valid species are known down there? Marine Biodiversity, 40 (3), 143-159 DOI: 10.1007/s12526-010-0041-4
Lambshead, P., & Boucher, G. (2003). Marine nematode deep-sea biodiversity – hyperdiverse or hype? Journal of Biogeography, 30 (4), 475-485 DOI: 10.1046/j.1365-2699.2003.00843.x




Why not spell the magic word money out? I’d rather say university bureaucracies and government bureaucracies have defunded taxonomy as a field.
“Nematodes are known from virtually every habitat in the seas, fresh water and on land. Some are generalists, but many have very specific habitats. One species is known only from felt coasters under beer mugs in a few towns in eastern Europe.”
Brusca, Richard C., and Gary J. Brusca. Invertebrates. Sunderland, MA: Sinauer Associates, 1990. Print.
I’ve never understood why taxonomy doesn’t get more funds. Look at the PR value COML have got recently for new species descriptions. AND, its one of the few fields where you are essentially guaranteed to get results. Its not like a multi-billion dollar particle collider that sits idle for 90% of the time. For a billion dollars, you could revolutionise taxonomy the world over! I’ve written about species accumulation curves before as a tool in predicting biodiversity. It seems to me that combining that approach with DNA barcoding we could at least make a huge dent in numerical diversity question without doing all the morphology. It would be straightforward and relatively easy, don’t you think? Not nearly as fund as new species descriptions, but maybe more realistic given the size of the task.
oops, I botched my HTML tags. MODS! sorry…
DNA barcoding does have one problem, and it’s a big one: it imposes one single species concept – and a phenetic one at that – on everyone.
To get that kind of very rough grip on nematode diversity, I’d prefer sequencing environmental DNA and running it through a phylogenetic analysis…
sorry, i posted oil in wrong site.
Taxonomy has always been on shaky ground, largely because its the aspect of the biological sciences that is more art than science… very few taxonomic papers would pass muster with scientists trained today, as they tend to be based on arbitrary and personal opinions on whether the differences between populations are sufficient to constitute new species (lumpers vs. splitters). Never mind how many species of nematodes there are, we can’t even say for sure how many species of field mice there are in North America, due to the wildly different opinions on whether or not two populations are sufficently differentiated to apply the species tag. With the ‘popular’ species it gets even worse… there are thousands of described cone snail species, but its estimated only about 600 are valid… all because different people went and described different extremes of the populations and separate species. In the less popular species you still have problems, as the species designations can all be down to one persons opinions, and there are no other experts to serve as a check on run-away imagination… in my own (former) career, I have encountered taxonomists who have built their careers on species designations (and publishing reams off biogeographic and evolutionary papers from these relationships) who have had everything overturned because someone else actually applied some statistics or genetics to show that their taxonomic trees were mere fantasy. DNA barcoding can help a bit with this situation, but on a highly variable and subdivided population it won’t tell you where the species boundaries actually are, but will increase the expense enormously.
Given these problems, is there any surprise that funding bodies shy away from funding taxonomy? When it was just a lone museum scientist surrounded by dusty cabinets and bottles of formaldehyde, it wasn’t that big an expense, and most funding came from patrons (victorian times) or core museum and university budgets. Now that access to a DNA lab is becoming the gold standard, questions are being raised about exactly how scientific much of taxonomy is, and the amount of money required to do the job properly has skyrocketed.