Lynn Margulis classified the Chaetognaths, known as arrow worms, as deuterostomes. Deuterostomy is characterized by several developmental characteristics including radial, indeterminate cleavage, a posterior position of the blastopore (deuterostomy=”second mouth”), enterocoelous coelom formation and a tripartite adult body plan with a post-anal tail. At least this is what I was taught “growing up”. Three papers published in Current Biology 2007 showed that while they have developmental patterns like deuterostomes, their genes tell another story!
Where Are All My Sistahs At?
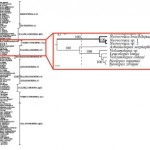
![]() The first paper by Matus et al. sampled several phyla using several genes. A consensus tree built from 72 EST (expressed sequence tags) genes grouped them within the Lophotrochozoa, which consists of molluscs and annelids among other phyla. They also constructed a tree using tropomyosin genes showing Chaetognaths as the sister taxon to all the Lophotrochozoa. One curiosity about the EST tree is that when they included the Priapulida, Chaetognaths grouped with them in the Ecdysozoa (Nematodes and Arthropods, among other phyla). When the data were reanalyzed without the Priapulids, Chaetognaths grouped with the last common ancestor of the molluscs and annelids. The authors argued that
The first paper by Matus et al. sampled several phyla using several genes. A consensus tree built from 72 EST (expressed sequence tags) genes grouped them within the Lophotrochozoa, which consists of molluscs and annelids among other phyla. They also constructed a tree using tropomyosin genes showing Chaetognaths as the sister taxon to all the Lophotrochozoa. One curiosity about the EST tree is that when they included the Priapulida, Chaetognaths grouped with them in the Ecdysozoa (Nematodes and Arthropods, among other phyla). When the data were reanalyzed without the Priapulids, Chaetognaths grouped with the last common ancestor of the molluscs and annelids. The authors argued that
“The association with the Ecdysozoa, therefore, depends on a single taxon and is not likely to reflect a general affinity for the group.”
But shouldn’t you use all the available evidence? Can you really justify leaving out data because it doesn’t fit your expectations? In their supplementary material, they show three trees with three different conclusions.
The first tree:
“Bayesian tree based on the analysis of RY-coded concatenated SSU and LSU rRNA genes shows a relationship between the chaetognaths Paraspadella and Sagitta with priapulids.”
Conclusion: Chaetognaths are sister-taxon to Priapulids, well situated within the Ecdysozoa.
The second tree:
“Bayseian analysis of Ribosomal SSU and LSU data, excluding priapulids from RY re-coded data… The maximum likelihood bootstrap tree had very low support values.”
Conclusion: Chaetognaths are sister-taxon to Platyhelminthes, well situated within the Lophotrochozoa.
The third tree:
“Bayesian analysis of Ribosomal SSU and LSU data excluding long branch platyhelminths and priapulids from RY re-coded data.”
Conclusion: Chaetognaths are sister-taxon to all the Lophotrochozoa, occupying a basal position to the clade. This is the tree they liked and went with for their main article.
Don’t get me wrong, I (think) I believe them. Their maximum likelihood studies show best support for Tree #3 and fits nicely into a story, keeps everything monophyletic and stable. For instance they state:
“To eliminate long branch attraction artefacts, we analyzed subsets of taxa to determine the effects of taxon sampling (Supplemental data). Regardless of the taxa removed (nematodes, platyhelminths or tardigrades) the chaetognaths remained as sister to annelids and molluscs with varying degrees of support in Bayesian analyses and as sister to the Ecdysozoa (arthropods, nematodes and tardigrades) with weak bootstrap support in likelihood analyses.”
There are profound evolutionary differences between protostomes and deuterostomes or between Lophotrochozoans and Ecdysozoans. Either way, the molecular data say Chaetognaths are definitely Protostomes. It disturbs me though that most of the developmental and morphological similarities to deuterostomes, as the authors stated, may be the result of convergent evolution. In my opinion, convergent evolution tends to destabilize tree topology by decreasing parsimony. Sometimes I feel that when you invoke convergent evolution too much you might as well through up your hands and proclaim the magnificence of the grand designer!
To Deuterostome or Not to Deuterostome, That is the Question
A complimentary study in the same issue of Current Biology by Marlétaz et al. consider the phylogeny of the Cheatognaths… again. Both studies analyzed genes of Chaetognaths and BLASTed the database of model organisms with those genes (mostly ESTs). The Marlétaz et al. paper actually did find something new and interesting though. They sequences over 11,000 ESTs of a juvenile Chaetognath (Spadella cephaloptera, below).

In their prodding around they found the gene for Guanidinoacetate N-methyltransferase. This gene is interesting because it is only found in Deuterostomes and Cnidarians, lost in the Protostomes. It catalyzes the final step towards creatine synthesis, which is an important compound in energy metabolism in brain and muscle tissue. Interesting that this is derived from a Cnidarian ancestor! Maybe that is why our brains look like jelly?
This provides striking evidence for their placement as a sister taxon to the Protostomes. Hence the common ancestor is not between a Protostome and Deuterostome, but between a Chaetognath and a deuterostome. May I remind you that Chaetognaths share the same embryological characteristics as Deuterostomes, plus now a Deuterostome-specific gene carried down from the Cnidarians and lost to the Protostomes. The phylogeny below from the Marlétaz et al. paper says it all.
 Problems? Well, like the Matus et al. paper, they exclude the Platyhelminthes from analysis. What is it about flatworms that confuses Chaetognath phylogeny?? For one, they were the previous occupants of the sister-clade-to-Protostomes position, otherwise known as the position between Cnidarians and Protostomes. Figure S2 from the online supplementary material shows the effects of this analysis. Fig. S2A includes Chaetognaths, while Fig S2B excludes them (below). As you can see, Platyhelminthes was excluded based on the weak bootstrap support either way. This begs the question about why are Chaetognaths and Platyhelminthes so closely related (potentially), while only Chaetognaths retain deuterostomy characteristics?
Problems? Well, like the Matus et al. paper, they exclude the Platyhelminthes from analysis. What is it about flatworms that confuses Chaetognath phylogeny?? For one, they were the previous occupants of the sister-clade-to-Protostomes position, otherwise known as the position between Cnidarians and Protostomes. Figure S2 from the online supplementary material shows the effects of this analysis. Fig. S2A includes Chaetognaths, while Fig S2B excludes them (below). As you can see, Platyhelminthes was excluded based on the weak bootstrap support either way. This begs the question about why are Chaetognaths and Platyhelminthes so closely related (potentially), while only Chaetognaths retain deuterostomy characteristics?
Márletaz et al. conclude:
“It confirms, on a genomic basis, that deuterostomy in an embryological sense is not a decisive character for the classification of animals. Some animals, like the chaetognaths, can be protostomes and yet show features of a deuterostome-like embryology. Nevertheless, the position of chaetognaths as a sistergroup of protostomes prompts us to propose that their development could be reminiscent of the bilaterian ancestor and testify that chaetognaths are a landmark phylum for addressing hypotheses about the origins of bilaterians.”
“Remarkable for the Obscurity of Their Affinities”

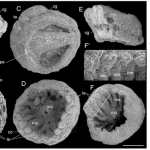
Yet again, in the same issue of Current Biology there exists a third article on Chaetognaths by Ball & Miller. This paper discusses more of the biology of chaetognaths and why they just plain cool creatures. They are a relic of the Cambrian and fossils show that not much has changed in their body plan (right).
“The chaetognaths are an ancient lineage of invertebrates that shares some characteristics with just about every other major invertebrate phylum and has consequently puzzled taxonomists ever since its original description in 1769. Darwin described chaetognaths as ‘‘remarkable for the obscurity of their affinities’’ and they have puzzled a succession of eminent zoologists ever since. Though unfamiliar to most biologists, chaetognaths are typically the most abundant planktonic predators, sometimes accounting for more than 10% of zooplankton biomass and being outnumbered only by their major prey, the copepods.”
 Chaetognaths feed on copepods by sensing their vibrations and are able hunters in darkness (click here for a movie of a chaetognath hunting!). Some even have a neurotoxic venom. They are also simultaneous hermaphrodites. One species, Paraspadella gotoi, exhibits interesting mating behavior with ritualistic dance before exchanging sperm packet (right).
Chaetognaths feed on copepods by sensing their vibrations and are able hunters in darkness (click here for a movie of a chaetognath hunting!). Some even have a neurotoxic venom. They are also simultaneous hermaphrodites. One species, Paraspadella gotoi, exhibits interesting mating behavior with ritualistic dance before exchanging sperm packet (right).
I found it interesting to that the authors note the nervous system bears a resemblance to nematodes, kinorhynchs and priapulids. In the phylogenetic papers of mentioned above, the authors take out priapulids from the analyses because they didn’t like that Chaetognaths grouped with them within the Ecdysozoa. The Ecdysozoa clade is supposed to encompass the moulting animals. So the Chaetognath’s nervous system resembles members of the Ecdysozoa AND Chaetognaths have a cuticle.
I have no doubt from the molecular work shown to date that chaetognaths align with protostomes. But the protostomes are a HUGE group encompassing some of the most diverse animal taxa such as the arthropods, nematodes, molluscs and annelids. Where they fit in that tree means quite a lot evolutionarily. There are many biologists that outright reject the Ecdysozoa hypothesis for a variety of reasons. Libbie Hyman, who wrote the book(s) on invertebrate diversity quite literally, recognized the affinities of the Chaetognaths to the protostomes:
‘‘It seems probable that the chaetognaths should be regarded as having diverged at an early stage from the primitive ancestor of the Bilateria.’’-Hyman quoted in Ball & Miller 2006

But Chaetognaths aren’t the only unresolved taxon. There are many minor taxa which have few people working on them. Some taxa don’t even have anyone in the U.S. as a specialist working on that group. Many of these taxa are in desperate need of study and revision and can potentially fill in several holes in animal phylogeny.
More Chaeognath Stuff:
Erik Thuesen’s Chaetognath Webpage
PZ Myers Chaetognath Friday Blogging (2004)
Ball, E., & Miller, D. (2006). Phylogeny: The Continuing Classificatory Conundrum of Chaetognaths Current Biology, 16 (15) DOI: 10.1016/j.cub.2006.07.006
Chen, J.-Y., & Huang, D.-Y. (2002). A Possible Lower Cambrian Chaetognath (Arrow Worm) Science, 298 (5591), 187-187 DOI: 10.1126/science.1075059
Marlétaz, F., Martin, E., Perez, Y., Papillon, D., Caubit, X., Lowe, C., Freeman, B., Fasano, L., Dossat, C., & Wincker, P. (2006). Chaetognath phylogenomics: a protostome with deuterostome-like development Current Biology, 16 (15) DOI: 10.1016/j.cub.2006.07.016
Matus, D., Copley, R., Dunn, C., Hejnol, A., Eccleston, H., Halanych, K., Martindale, M., & Telford, M. (2006). Broad taxon and gene sampling indicate that chaetognaths are protostomes Current Biology, 16 (15) DOI: 10.1016/j.cub.2006.07.017








I remember taking inverts back in ’99. The papers that first proposed the Lophotrochozoan/Ecdysozoan split were just really gaining traction, although the textbooks hadn’t been revised yet. For my class project, I wrote them up and explained the new phylogeny as opposed to the one we were being taught. I thought it was pretty cool stuff, and often chuckled at the old/new split.
The final exam for the class was to draw the tree of invertebrate life down to the class level. So, I went up to my prof and asked him which I should do. He just laughed, called me a smartass, and told me to write up whichever.
(Also, the whole acoel thing still blows my mind.)
I have to admit I’ve about given up on invert phylogeny and systematics. I’m so out of date after only a few years not paying attention to it, and it seems a hopeless web. None of these papers are considering the potential for lateral gene transfer, I suppose some entire phyla could have some funky origins if Mike Arnold is right….
I don’t know about the ESTs but think about all the phylogenies out there based on single or a few markers, like COI or 16s, 18s, 28S, etc… the usual suspects in invert phylogeny. It is hard to keep in mind when doing molecular phylogenies that one is really tracking the gene history (I’m guilty too!), not the species history. To really do a good phylogeny, you need to make a case using morphology, fossils, select genes (or the genome), and maybe some behavior/ecology. Then you are still only guessing, but the argument is much stronger. Lateral gene transfer really puts a wrench in things and with recent, exciting research it seems to be a much bigger player in eukaryotes. I find it exciting though, maybe my research will take me there in one shape or another.
Wow, what an informative article! I’m working on a series of articles about animal phyla and used your article as a reference for my Chaetognatha page at http://www.geozoo.org/Life/Chaetognatha I included a link to your page.
I’ll have to check out some more articles on your site. Thanks!