![]() …because “High-throughput sequencing confers a deep view of seasonal community dynamics in pelagic marine environments”, however appropriate a title, seems far too dry and technical for a blog. I mean, I want people to read my posts, right? Don’t be fooled by the title, though: I am going to give you some seriously elegant science here.
…because “High-throughput sequencing confers a deep view of seasonal community dynamics in pelagic marine environments”, however appropriate a title, seems far too dry and technical for a blog. I mean, I want people to read my posts, right? Don’t be fooled by the title, though: I am going to give you some seriously elegant science here.
Because of my own research, I try to keep pretty up-to-date with the wider world of high-throughput DNA sequencing—keeping tabs on other scientists who, like myself, use millions of DNA sequences to study complex species assemblages in different types of ecosystems. A couple recent papers relayed results from a 6-year investigation of microbial communities in the Western English Channel waters near Plymouth, UK.
Gilbert et al. (2011, 2010) show that even in bacterial communities, there are definite seasonal patterns and peaks in community diversity. Figuring out what causes these patterns is sometimes surprisingly easy – it looks like shifting day length accounts for 65% of the changes in bacterial diversity (I’m sure the authors’ jaws dropped when they saw this result…). Even more ridiculous (in a good way), the specific bacterial assemblage—the ‘fingerprint’ of species present in the community—could predict the month with 100% accuracy. And no surprise, only 2% of the 100 most abundant taxa they observed could be identified down to species level. (Previously undiscovered diversity is so old hat these days. But still cool).
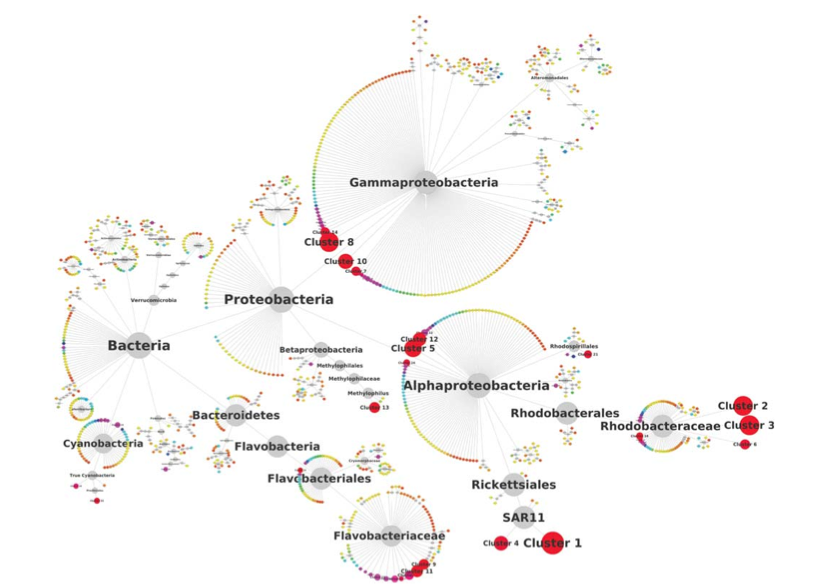
Network analysis indicates that communities are driven from the “bottom-up”: the environment determines the bacterial taxa that thrive (differential responses to temperature, light availability, etc), and then the eukaryotic taxa may then sneak in depending on what yummy bacteria there are to eat:
“Reginald, I do fancy some Gammaproteobacteria for supper on this fine afternoon – what do you say”
“Tickety-boo old chap, lets get swimming”
Over time there were occasional blooms of rare taxa – all of a sudden one species would become perplexingly abundant. These types of spikes are very exciting – as a scientist, I dream of the day where we can link a specific taxon to a specific environmental change (invasion of predators, an oil spill, or an invisible pollutant). As more studies like these are published, my dream may soon become a reality.

The coolest thing is that these patterns are consistent no matter what type of data you look at (Gilbert et al. 2010) – whether you’re looking at one gene sequenced from all species (metagenetics), random genomic fragments from the community (metagenomics), or only the community gene expression (metatranscritpomics). From gene expression data, Gilbert et al. additionally found important genes which turn on and off according to season and time of day – but these show NO resemblance to any known genes. Which is pretty exciting, because we’ve sequenced a lot of genomes at this point, and functionally important genes are pretty conserved (and easy to spot) across the tree of life. Microbes are obviously doing something important in the environment—maybe producing an unknown protein which recycles nutrients such as Nitrogen or Phosphorus—but all we can do is scratch our heads at this point. New studies like this are constantly turning the science world upside down—like, we scientists thought we knew how the Nitrogen cycle worked…but apparently we didn’t.
Although Gilbert et al. (2011) focused on UK waters, their observations of seasonal patterns were surprisingly congruent with a previous study off the California coast (Fuhrman et al. 2006). To me, the fact that microbial communities can be so similar, despite being so geographically far apart, is completely amazing. But like any good scientific study, the results seem to generate more questions than answers. Now that we’re building a deep and accurate view of marine ecosystems (thank you, high-throughput sequencing), we can start to dig further and ask questions about what exactly drives these patterns. I can’t wait to find out!
References:
Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrman JA, & Field D (2011a). Defining seasonal marine microbial community dynamics. The ISME journal PMID: 21850055
Gilbert, J., Field, D., Swift, P., Thomas, S., Cummings, D., Temperton, B., Weynberg, K., Huse, S., Hughes, M., Joint, I., Somerfield, P., & Mühling, M. (2010). The Taxonomic and Functional Diversity of Microbes at a Temperate Coastal Site: A ‘Multi-Omic’ Study of Seasonal and Diel Temporal Variation PLoS ONE, 5 (11) DOI: 10.1371/journal.pone.0015545
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, & Naeem S (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proceedings of the National Academy of Sciences of the United States of America, 103 (35), 13104-9 PMID: 16938845




One Reply to “Big text files can tell you how the ocean works”
Comments are closed.